Recently, Prof. Jianyang Zeng’s research group from Institute for Interdisciplinary Information Sciences (IIIS), Tsinghua University, published their latest work in Cell Systems (a sister journal of Cell), entitled "Analysis of Ribosome Stalling and Translation Elongation Dynamics by Deep Learning". This paper describes the first application of deep learning technology to the modeling of protein translation dynamics. An early version of this paper was accepted previously by the 21st International Conference on Research in Computational Molecular Biology (RECOMB 2017) as an oral presentation in the ‘New Direction’ Session. Overall, it presents a new deep learning based framework facilitated by the high-throughput sequencing technology, and provides useful insights into revealing the basic regulatory mechanisms underlying protein translation.
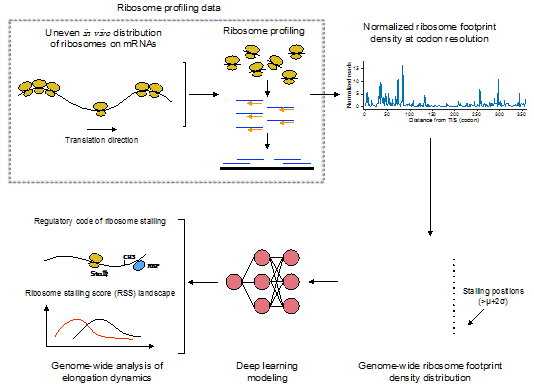
Protein translation is the key process in the delivery of genetic information from DNA to protein, as described in Central dogma of molecular biology. In protein translation, an organelle, called ribosome, scans of the messenger RNA (mRNA) chain and translates the nucleotide sequence into a corresponding amino acid sequence, specified by genetic code. In recent years, through the development of high-throughput sequencing technology, biologists have found the process of protein translation to be highly dynamic. In particular, a phenomenon called ribosome stalling occurs from time to time, which is closely related to protein folding and localization.
As a breakthrough in this field, Prof. Jianyang Zeng’s Research Group introduced the machine learning techniques to the mechanical and functional studies of ribosome stalling. In particular, they successfully developed a deep learning based framework for predicting ribosome stalling. This model only relies on the primary sequence mRNA as input, i.e., it does not explicitly integrate other biological features. Furthermore, based on the prediction results, they systematically analyzed various putative factors regulating ribosome stalling and how ribosome stalling is associated with biological functions. Meanwhile, they proposed many new hypotheses, in order to lay a solid foundation for the further exploration of protein translation dynamics.
International Conference on Research in Computational Molecular Biology (RECOMB) is one of the two top conferences in the field of computational biology, with an average annual acceptance rate of about 20%. Sai Zhang (PhD student), Hailin Hu (PhD student) and Jingtian Zhou (undergraduate student) are the co-first authors of this work, with Prof. Zeng the corresponding author. This is a collaborating work with Prof. Tao Jiang from University of California, Riverside. Cell Systems is an uprising interdisciplinary journal from Cell Press. It publishes high-profile articles on ‘systems-level understanding in the life sciences’. As a sister journal of the world's top biology journal Cell, Cell System has achieved great influence all over the world after its first two-year publication.
The full paper is available at:
http://www.cell.com/cell-systems/fulltext/S2405-4712(17)30337-X
https://doi.org/10.1093/bioinformatics/btx247