Three-dimensional chromosome structure modeling is crucial to understanding how the faraway regulatory elements on the genomic map come close to each other in the 3D space. Both Hi-C and FISH are two popular techniques for inspecting chromosomes. Through contact frequencies, Hi-C gives an indirect way to investigate how two genomic loci are far or close to each other in the 3D space. On the other hand, FISH data can directly provide the average spatial distance between two genomic loci through imaging them.
Prof. Jianyang Zeng‘s group at the Institute for Interdisciplinary Information Sciences (IIIS) and collaborators, proposed GEM-FISH, a method that systematically integrates both Hi-C and FISH data with the available prior biophysical knowledge about 3D polymers to reconstruct the 3D chromosome models. The work is recently published in Nature Communications. In the paper, they have shown that GEM-FISH can reconstruct more accurate 3D chromosome models than when using Hi-C data alone in terms of the accurate positioning of topologically associated domains (TADs) and their partitioning into the two compartments A and B. In addition, GEM-FISH could accurately capture the high-resolution features of the chromosomes. For instance, it could capture the spatial proximity of loop loci, the colocalization of genomic loci from the same subcompartment, the tendency of expressed genes to lie close to the chromosome surface. They have also found interesting patterns of the spatial distributions of super-enhancers on the three investigated autosomes (Chrs 20, 21, and 22), which can provide useful insights on the regulatory roles of super-enhancers in controlling the expression of cell identity genes.
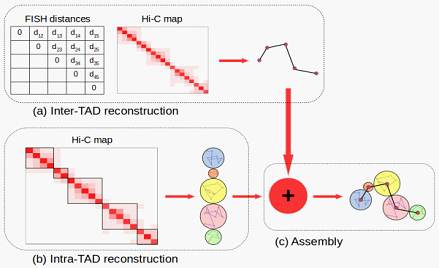
Flowchart of the new 3D genome structure modeling appraoch
This work is joint work with Prof. Michael Zhang (Department of Biological Sciences, Center for Systems Biology, the University of Texas at Dallas and MOE Key Laboratory of Bioinformatics, Tsinghua University) and Prof. Juntao Gao (MOE Key Laboratory of Bioinformatics, Tsinghua University). Prof. Jianyang Zeng, leader of the Machine Learning and Computational Biology research group at the Institute for Interdisciplinary Information Sciences (IIIS) at Tsinghua University, and his post-doc Dr. Ahmed Abbas are the corresponding and first authors of the paper, respectively. The work is also supported by National Natural Science Foundation of China.
The full paper is available at https://www.nature.com/articles/s41467-019-10005-6.