The research team of Jianyang Zeng, IIIS Associate Professor, has made a pioneering breakthrough of a novel manifold learning based framework, called GEM (Genomic organization reconstructor based on conformational Energy and Manifold learning), to reconstruct the three-dimensional organizations of chromosomes by integrating Hi-C data with biophysical feasibility. The research is published in Nucleic Acids Research, and recognized in Top Ten Advances in Bioinformatics of China 2018 recognized by the peer reviewers organized by Genomics, Proteomics and Bioinformatics (GPB) journal.
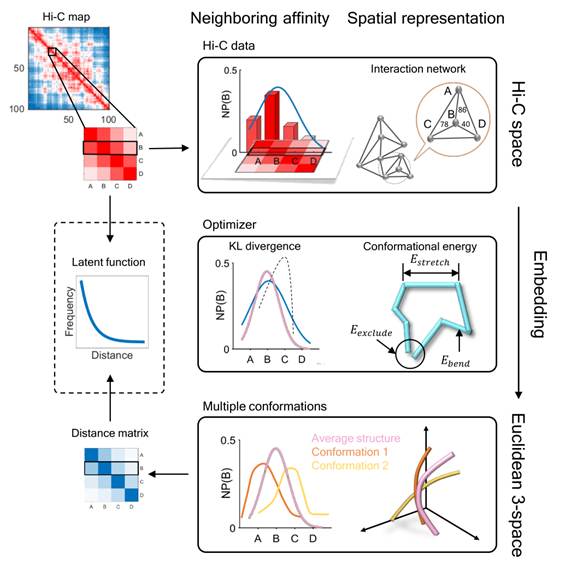
The 3D packaging of the genome in the nucleus plays an important role in many cellular processes, including DNA replication, transcription, and DNA repair. Reconstruction of the 3D genomic architecture can provide important implications into understanding the underlying mechanisms of gene activities. Recently, chromosomal conformation capture based technologies, such as Hi-C, have been used to investigate the 3D folding the genome. Dr. Jianyang Zeng’s team has recently developed a novel manifold learning based framework, called GEM, to reconstruct the 3D organizations of chromosomes by integrating Hi-C data with biophysical energy. Unlike previous methods, it directly embeds the neighboring affinities from Hi-C space into 3D Euclidean space. Moreover, Dr. Zeng’s team for the first time apply the modeled chromatin structures to recover long-range genomic interactions missing from original Hi-C data.
The work was co-conducted by Prof. Tommy Kaplan from Hebrew University of Jerusalem, and Prof. Jian Peng from University of Illinois at Urbana–Champaign. Guangxiang Zhu, IIIS PhD candidate is the first author of this paper and Prof. Jianyang Zeng is the correspondence author. The work was also supported by National Natural Science Foundation of China and Beijing Advanced Innovation Center for Structural Biology.
The full paper is available at https://academic.oup.com/nar/article/46/8/e50/4835049