Recently, Prof. Jianyang Zeng’s research group from Institute for Interdisciplinary Information Sciences (IIIS), Tsinghua University, published their latest work in Proceedings of the National Academy of Sciences of the United States of America (PNAS), entitled "A machine learning based framework for modeling transcription elongation". This paper demonstrates the first deep learning based model for predicting RNA polymerase II (Pol II) pausing sites.
During the transcription elongation, Pol II may pause in specific regions and accumulate on different sites of gene bodies, named Pol II pausing. Plenty of studies have demonstrated that Pol II pausing is often coupled with co-transcriptional events, such as RNA processing, modulating transcription elongations rates, gene expression regulation and alternative splicing. Native elongating transcript sequencing (NET-seq) is a new technique that can detect the whole genome Pol II pausing sites at strand-specific and single nucleotide resolution. However, the traditional sequencing techniques is generally time- and resource-consuming. In addition, the sequence features of Pol II pausing and the relations between Pol II pausing and regulation of transcription elongation have not been fully studied. Prof. Jianyang Zeng’s research group introduced a machine learning framework to comprehensively study the mechanisms of Pol II pausing. The models only takes the primary DNA sequence as input to predict the Pol II pausing sites and achieves a superior performance compared with the state-of-the-art methods. Furthermore, by analyzing the attention mechanism employed in the model and the prediction results on specific biological sites, they systematically demonstrated the underlying sequence features of the Pol II pausing and the relations between Pol II pausing and transcription factors, histone modifications, DNA methylations and types of alternative splicing. The framework provides a new computational method to complement the low sequencing depth of current techniques and useful understandings of co-operations between Pol II pausing and transcription elongation events.
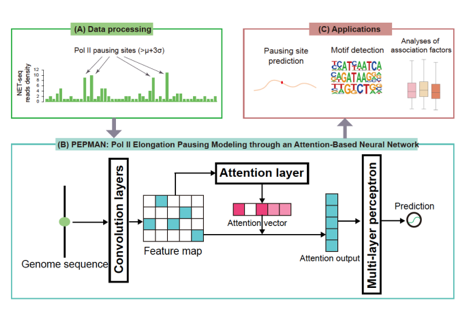
An overview of RNA polymerase II pausing modeling framwork
Peiyuan Feng (PhD student) and An Xiao (Master student) are the co-first authors of this work, and Prof. Zeng and Dr. Dan Zhao are the corresponding authors. This work is supported by National Natural Science Foundation of China, Turing AI Institute of Nanjing and Zhongguancun Haihua Institute for Frontier Information Technology.
(See the link for the published paper https://www.pnas.org/content/118/6/e2007450118 )